Isotope protein labeling
Isotope protein labeling
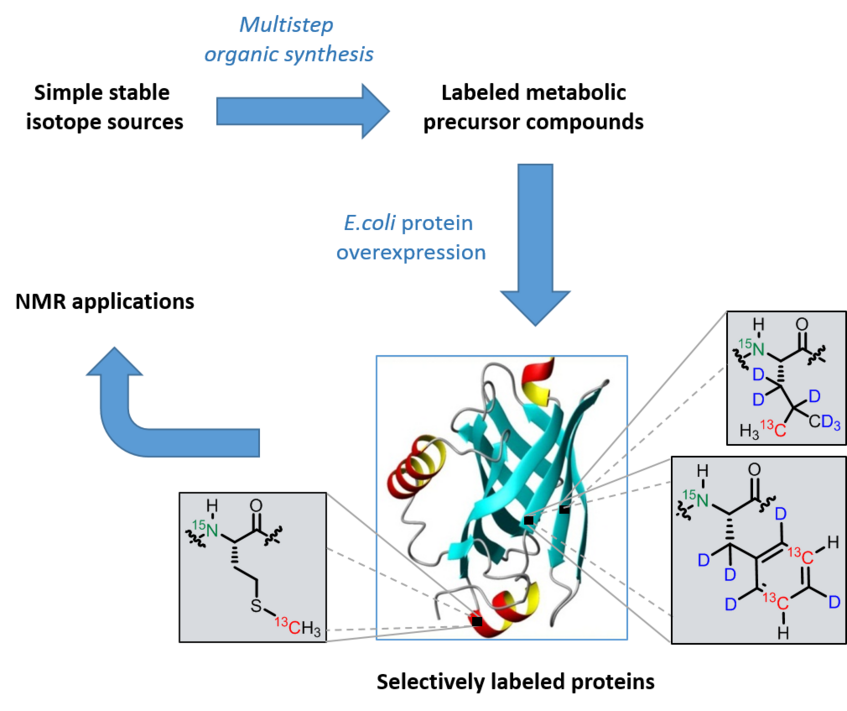
The rapid development of biomolecular NMR spectroscopy in the last decade(s) has paved the way for novel insights into the structure, dynamic properties, as well as the interaction of proteins. However, NMR data interpretation of large protein complexes is only feasible if the corresponding sample is selectively enriched in NMR active nuclei (13C and 15N) and/or 1H-depleted by 2H incorporation. One important strategy for selective protein isotope labeling is given by the addition of metabolic amino acid precursor compounds to the growth medium of a protein-overexpressing microorganism.
In our current projects, we investigate robust synthetic routes from cheap isotope sources to selectively labeled precursors. These compounds show an effective uptake into the metabolism of the overexpressing E. coli strains. Our techniques result in well defined 13C, 15N and 2H patterns in the corresponding target amino acids devoid of any cross labeling to other residues. Our proteins can be used for various biomolecular NMR experiments: They can provide additional restraints for structure calculation, simplify NMR spectra derived from large protein complexes, exhibit isolated spin systems for accurate interpretation of dynamic processes and offer well defined signals for straightforward chemical shift mapping.
Visit the Gallery of precursor compounds to see a selection of small molecules, which can be used for selective labeling of Ile, Leu, Val, Phe, Tyr, Trp or His residues.
Pi by NMR
Pi by NMR
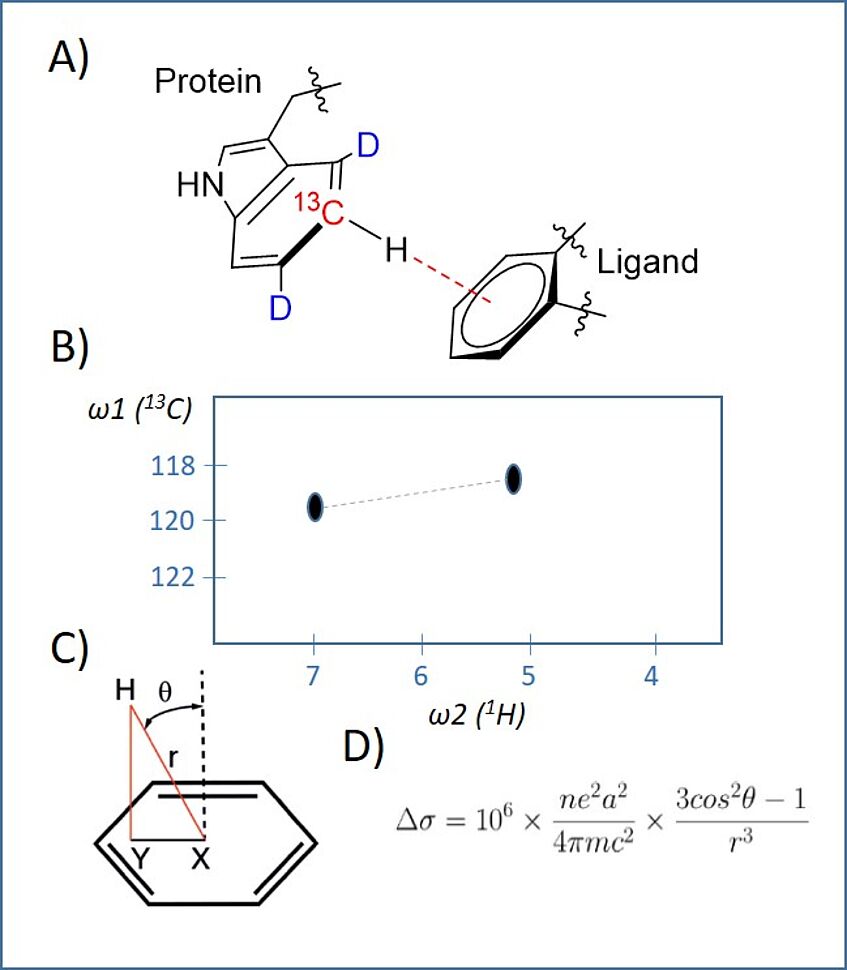
The Pi system of aromatic rings serves as an important source of non-covalent interaction between drug molecules and their corresponding target proteins. In addition to arene-arene and ion-π interaction, hydrogen bonding to π-surfaces acts as a major modulator of affinity and selectivity in protein-ligand complexes. NMR spectroscopy is a powerful method to collect valuable data on site-specific interaction at atomic resolution. However, suitable experiments to quantify CH-π interactions have not been reported, yet. Together with the group of Robert Konrat at the Max-Perutz Laboratories (University of Vienna) and Boehringer-Ingelheim as an industry partner, we combined selective labeling and sensitive 1H-13C NMR spectroscopy to decipher the details of CH-π binding. We applied our synthetic protocol to access isotopologues of anthranilic acid, showing 13C-H spin systems surrounded by 12C-D. These compounds have been applied in E. coli based protein overexpression, which resulted in highly selective Trp labeling in the target proteins. The Trp 13C-H resonances experience a significant chemical shift perturbation upon ligand interaction (A and B) in HSQC spectra. A set of ligands with known protein-ligand structures was analyzed. Using these data sets, the change in isotropic nuclear shielding constant (Δσ) could be correlated to the distance between the proton to the ring center (r) and the angle between the ring normal and the proton position (ϑ) via the Pople equation (C and D). Pi by NMR is a new potent tool to specifically optimize CH-π interactions in the design process of molecular therapeutics.
This work was supported by the Christian Doppler Laboratory for Knowledge-based Structural Biology and Biotechnology. Results have been published in Angew. Chem. Int. Ed. 2020, 59, 35, 14861-14868 (open access).
